FROM SCRIBBLES TO STRUCTURED DATA: PROCESSING HANDWRITTEN PRESCRIPTIONS WITH SPARK NLP

Introduction
Medical prescriptions, often scribbled in hurried handwriting, pose a significant challenge when attempting to extract valuable information.
Automating this process requires a robust combination of Optical Character Recognition (OCR) and Natural Language Processing (NLP) tools to accurately identify entities like medication names, dosages, and medical conditions.
In this article, we delve into a Spark NLP-based pipeline to convert handwritten prescriptions into structured, machine-readable text. Leveraging BERT embeddings for contextual understanding and a custom Named Entity Recognition(NER) model, this approach promises to streamline information extraction in medical workflows. From OCR text extraction to entity recognition and model training, each step is tailored to maximize accuracy for complex medical terminology.
Table of Contents
- 1. Extract Handwritten text using OCR
- 2. Initializing the NER Model
- 3. Training the Model
- 4. Loading the Model
- 5. Making Predictions
- What’s a Document?
- Summary
- Actual Output
- Conclusion
- References
1. Extract Handwritten text using OCR
The first and foremost step is to use an OCR to extract handwritten texts from the doctor’s prescriptions. I have used AWS Textract that automatically extracts text, handwriting, layout elements, and data from scanned documents.
def detect_text(local_file, region_name, aws_access_key_id, aws_secret_access_key):
# Initialize Textract client
textract = boto3.client(
'textract',
region_name=region_name,
aws_access_key_id=aws_access_key_id,
aws_secret_access_key=aws_secret_access_key
)
# Open file and detect text
with open(local_file, 'rb') as document:
response = textract.detect_document_text(Document={'Bytes': document.read()})
# Extract text from response
text_lines = [
item["Text"]
for item in response.get("Blocks", [])
if item.get("BlockType") == "LINE"
]
extracted_text = " ".join(text_lines)
logger.info(f"Successfully extracted text from {local_file}")
return extracted_text
The data extracted from the OCR is then sent further into the NER pipeline, which we will be creating later in this blog, for extraction of Named Entities.
2. Initializing the NER Model
We create a class InitiateNER to initialize the Spark NLP session, embeddings, and NER tagger.
class InitiateNER:
def __init__(self, gpu=True, embedding_model='bert_base_uncased', language='en'):
# Initialize Spark session
self.spark = sparknlp.start(gpu=gpu)
logger.info("Spark NLP session started.")
# Initialize embeddings and NER tagger
self.bert_embeddings = BertEmbeddings.pretrained(embedding_model, language) \
.setInputCols(["sentence", 'token']) \
.setOutputCol("embeddings") \
.setCaseSensitive(False)
self.ner_tagger = NerDLApproach() \
.setInputCols(["sentence", "token", "embeddings"]) \
.setLabelColumn("label") \
.setOutputCol("ner") \
.setMaxEpochs(20) \
.setLr(0.001) \
.setPo(0.005) \
.setBatchSize(32) \
.setValidationSplit(0.1) \
.setUseBestModel(True) \
.setEnableOutputLogs(True)
# Initialize other components for prediction
self.document_assembler = DocumentAssembler() \
.setInputCol("text") \
.setOutputCol("document")
self.sentence_detector = SentenceDetector() \
.setInputCols(['document']) \
.setOutputCol('sentence')
self.tokenizer = Tokenizer() \
.setInputCols(['sentence']) \
.setOutputCol('token')
self.converter = NerConverter() \
.setInputCols(["document", "token", "ner"]) \
.setOutputCol("ner_span")
The NerDLModel is an encoder-decoder neural network (we will talk about it later) which needs the input in the form of embeddings[1]. This can be achieved via a WordEmbeddings model. We are using BertEmbeddings in this case.
WordEmbeddings model
Word embeddings[2] capture semantic relationships between words, allowing models to understand and represent words in a continuous vector space where similar words are close to each other. This semantic representation enables more nuanced understanding of language.
Why I chose BERT Embeddings over others?
BERT embeddings [3, 4, 5, 6] offer several advantages over traditional embeddings for Named Entity Recognition (NER):
-
Contextualized Embeddings: BERT creates different embeddings for the same word based on its context, unlike Word2Vec or GloVe, which give each word a single fixed representation. This is crucial in NER, where the meaning of a word can change with context.
-
Bidirectional Attention: BERT reads sentences in both directions (left-to-right and right-to-left), capturing a full understanding of each word’s context. Traditional embeddings lack this bidirectional context, missing nuances especially useful in medical text.
-
Pre-Trained on Large Data: BERT is pre-trained on a large corpus, which enhances its versatility for fine-tuning in domain-specific applications like medical NER, where context-rich understanding is needed.
-
Handling Rare or OOV Words: BERT uses subword tokenization, which allows it to break down unknown words and still retain meaning, a huge advantage for handling medical terminology.
-
Proven Performance: BERT consistently outperforms older embeddings on NER tasks due to its deep contextual understanding and flexibility in handling diverse language structures.
NerDLApproach
The NerDLApproach[1] in Spark NLP is a neural network model designed for Named Entity Recognition (NER) tasks. It uses a combination of Character-level Convolutional Neural Networks (Char CNNs), Bidirectional Long Short-Term Memory networks (BiLSTMs) (Char CNN - BiLSTM[7]), and followed by a Conditional Random Fields (CRFs). Let’s break down why this architecture is used and how it processes normal text input.
Why Char CNNs - BiLSTM - CRF?
-
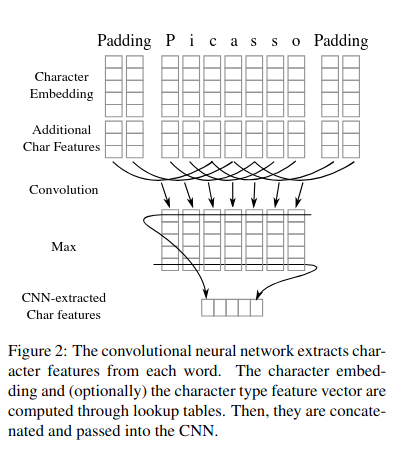
Character-level CNNs (Char CNNs):
-
Purpose: Capture morphological features of words, such as prefixes, suffixes, and other subword patterns.
-
Input: Character embeddings of each word.
-
Output: Character-level features that are useful for understanding the structure of words, especially for handling out-of-vocabulary words or misspellings.
-
-
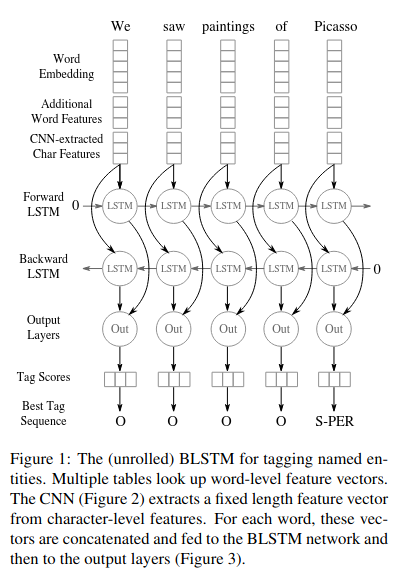
Bidirectional LSTMs (BiLSTMs):
-
Purpose: Capture contextual information from both past (left) and future (right) contexts in the text.
-
Input: Word embeddings (e.g., BERT embeddings) and character-level features from Char CNNs.
-
Output: Contextualized word representations that consider the entire sentence.
-
-
Conditional Random Fields (CRFs):
- Purpose: Model the dependencies between output labels (e.g., the sequence of named entity tags) to ensure valid sequences.
- Input: Contextualized word representations from BiLSTMs.
- Output: The most likely sequence of named entity tags for the input text.
Example
Let’s consider an example to illustrate this process:
Input Text: "John Doe works at Store Ninja."
-
Tokenization:
- Tokens:
["John", "Doe", "works", "at", "Store", "Ninja", "."]
- Tokens:
-
Character Embeddings:
- Characters for “John”:
["J", "o", "h", "n"] - Character embeddings are generated for each character.
- Characters for “John”:
-
Char CNNs:
- Char CNNs process the character embeddings to capture morphological features.
-
Word Embeddings:
- Word embeddings (e.g., BERT embeddings) are generated for each token.
-
BiLSTMs:
- BiLSTMs process the word embeddings and character-level features to capture contextual information.
- For example, the representation for “John” will consider the context provided by “Doe works at Store Ninja.”
-
CRFs:
- CRFs predict the sequence of named entity tags.
- Output:
["B-PER", "I-PER", "O", "O", "B-ORG", "O"]
3. Training the Model
The train_model method reads the CoNLL dataset, applies BERT embeddings, and trains the NER model.
def train_model(self, training_file, save_path):
try:
training_data = CoNLL().readDataset(self.spark, training_file)
training_data = self.bert_embeddings.transform(training_data).drop("text", "document", "pos")
logger.info("Training model...")
self.ner_model = self.ner_tagger.fit(training_data)
logger.info("Model trained successfully.")
self.ner_model.write().overwrite().save(save_path)
logger.info(f"Model saved at: {save_path}")
except Exception as e:
logger.error(f"Error during training: {e}")
train_modelmethod: Trains the NER model.CoNLL().readDataset: Reads training data in CoNLL format.self.bert_embeddings.transform: Applies BERT embeddings to the training data.self.ner_tagger.fit: Trains the NER model.self.ner_model.write().overwrite().save: Saves the trained model.
What is CoNLL format?
The CoNLL (Conference on Natural Language Learning) format is a structured text format commonly used to label tokens in natural language processing tasks, especially named entity recognition (NER). In a CoNLL-formatted dataset, each line represents a token (such as a word or punctuation), with additional columns typically containing labels or annotations about that token. Sentences are separated by blank lines.
CoNLL Format Structure
A typical CoNLL dataset has multiple columns for each token. The columns can vary based on the dataset’s intended task, but for NER, they usually look like this:
Token POS Chunk Entity
John NNP B-NP B-PER
lives VBZ B-VP O
in IN B-PP O
New NNP B-NP B-LOC
York NNP -X- I-LOC
. . O O
In this example:
- Each line corresponds to a word, and there may be additional columns with part-of-speech (POS) or chunk tags.
- The “Entity” column marks NER tags (e.g., B-PER for “Beginning of a PERSON entity” and O for “Outside of any entity”).
- Blank lines separate sentences.
- -X- represents that is often used as a placeholder or dummy value for certain columns, especially when the information in that column is not relevant for a particular token.
The dataset I am using looks something like this -
Token POS Chunk Entity
negative -X- -X- O
for -X- -X- O
chest -X- -X- B-Medical_condition
pain, -X- -X- I-Medical_condition
pressure, -X- -X- B-Medical_condition
dyspnea, -X- -X- B-Medical_condition
edema -X- -X- B-Medical_condition
or -X- -X- O
cold -X- -X- B-Medical_condition
symptoms. -X- -X- O
As we are only interested in Entity mapping, the POS and Chunk values are irrelevant to us, hence we have put a -X- or Empty Placeholder there.
Why CoNLL Format Is Used in NER
The CoNLL format is widely used in NER for several reasons:
- Structured yet simple: It provides a structured, line-by-line format that’s easy to parse and interpret, both by humans and machines.
- Consistent token-label pairing: Each token is directly paired with its label, simplifying the process of supervised learning where the model learns to associate tokens with specific tags.
- Sentence separation: Blank lines make sentence boundaries clear, which is crucial for tasks like NER, where context within sentences matters.
- Widely supported by NLP tools: Many NLP frameworks (like spaCy, NLTK, and transformers libraries) support CoNLL format, so training data in this format is easily integrated into NER pipelines.
Where to get Clinical NER Datasets?
Such datasets can be found at various sources -
- CoNLL-2003[8]
- 2010 i2b2/VA[9]
- 2014 i2b2 De-identification Challenge
- 2018 n2c2 Medication Extraction Challenge
4. Loading the Model
def load_model(self, model_path):
try:
self.loaded_ner_model = NerDLModel.load(model_path) \
.setInputCols(["sentence", "token", "embeddings"]) \
.setOutputCol("ner")
logger.info("Model loaded successfully.")
except Exception as e:
logger.error(f"Error loading model: {e}")
load_modelmethod: Loads a pre-trained NER model.NerDLModel.load: Loads the model from the specified path.
5. Making Predictions
def predict(self, text):
try:
# Create or reuse prediction pipeline
if not hasattr(self, 'prediction_pipeline'):
self.prediction_pipeline = Pipeline(
stages=[
self.document_assembler,
self.sentence_detector,
self.tokenizer,
self.bert_embeddings,
self.loaded_ner_model,
self.converter
]
)
logger.info("Prediction pipeline created.")
sample_data = self.spark.createDataFrame([[text]]).toDF("text")
prediction_model = self.prediction_pipeline.fit(self.spark.createDataFrame([['']]).toDF("text"))
preds = prediction_model.transform(sample_data)
pipeline_result = preds.collect()[0]
return self.format_output(pipeline_result)
except Exception as e:
logger.error(f"Error during prediction: {e}")
return {"error": str(e)}
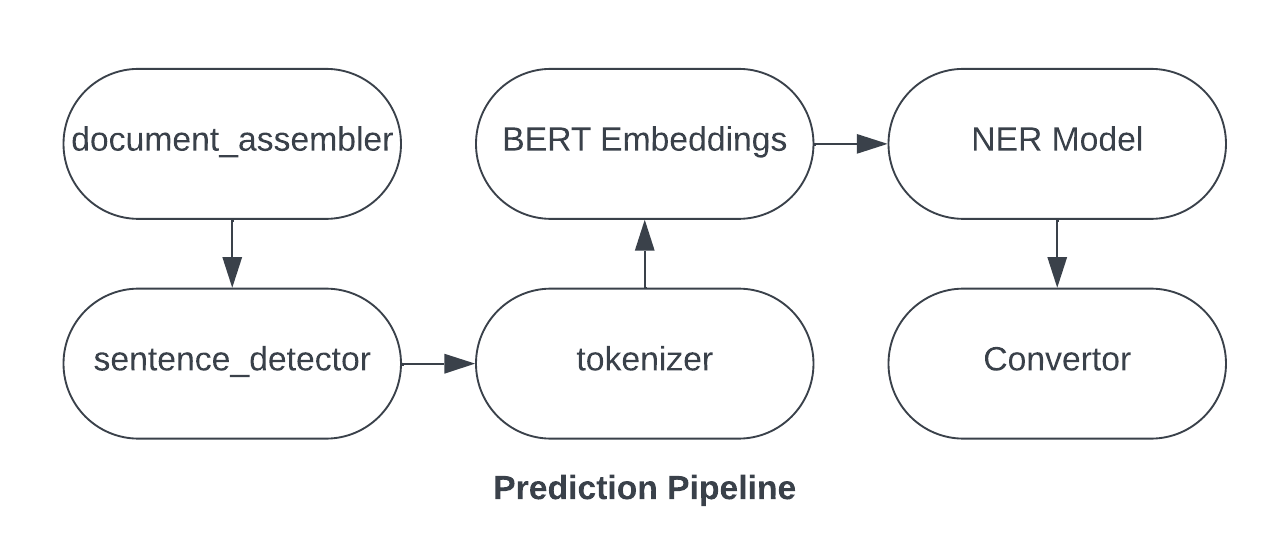
-
Pipeline Stages: The pipeline consists of several stages:
document_assembler: Converts raw text into a structured document format (read about it below).sentence_detector: Detects sentences within the document.tokenizer: Tokenizes sentences into individual words.bert_embeddings: Applies BERT embeddings to the tokens.loaded_ner_model: Uses the pre-trained NER model to identify named entities.converter: Converts the NER results into a more readable format.
-
Pipeline Example
-
If the input text is
"John Doe works at OpenAI.", the pipeline stages will process it as follows:- DocumentAssembler: Converts the text into a document.
- SentenceDetector: Identifies
"John Doe works at OpenAI."as a single sentence. - Tokenizer: Tokenizes the sentence into
["John", "Doe", "works", "at", "OpenAI", "."]. - BertEmbeddings: Generates embeddings for each token.
- Loaded NER Model: Identifies
"John Doe"as a person and"OpenAI"as an organization. - Converter: Converts the NER results into a readable format.
-
-
self.spark.createDataFrame: Creates a Spark DataFrame from the input text. -
self.prediction_pipeline.fit: Fits the pipeline (required for some Spark operations). -
prediction_model.transform: Transforms the input text using the pipeline. -
preds.collect: Collects the prediction results. -
self.format_output: Formats the prediction results.
What’s a Document?
In the context of Natural Language Processing (NLP) and Spark NLP, a “document” refers to a structured representation of text data. This structured representation is used as an intermediate format that allows various NLP components to process the text more effectively. Let’s break down what this means in more detail:
Document in Spark NLP
A “document” in Spark NLP is essentially a DataFrame column that contains metadata about the text, such as its content, the start and end positions of the text, and other relevant information. This structured format is crucial for the subsequent NLP tasks, such as sentence detection, tokenization, and named entity recognition.
DocumentAssembler
The DocumentAssembler[10] is a component in Spark NLP that converts raw text into this structured “document” format. Here’s how it works:
- Input Column: The
DocumentAssemblertakes a column of raw text as input. - Output Column: It produces a column of “document” type, which contains the structured representation of the text.
Example
Let’s look at an example to understand this better:
from sparknlp.base import DocumentAssembler
from pyspark.sql import SparkSession
# Initialize Spark session
spark = SparkSession.builder \
.appName("DocumentAssemblerExample") \
.getOrCreate()
# Sample DataFrame with raw text
data = spark.createDataFrame([["This is a sample text."]]).toDF("text")
# Initialize DocumentAssembler
document_assembler = DocumentAssembler() \
.setInputCol("text") \
.setOutputCol("document")
# Transform the DataFrame
document_df = document_assembler.transform(data)
# Show the result
document_df.select("document").show(truncate=False)
Output
The output will be a DataFrame with a “document” column that contains the structured representation of the text:
+-------------------------------------------------------------+
|document |
+-------------------------------------------------------------+
|[[document, 0, 20, This is a sample text., [sentence -> 0], []]]|
+-------------------------------------------------------------+
Explanation
document: The column name for the structured text.0, 20: The start and end positions of the text.This is a sample text.: The actual text content.[sentence -> 0]: Metadata indicating that this is the first sentence.
Why is this Important?
The structured “document” format is essential for the following reasons:
- Consistency: It provides a consistent way to represent text data, making it easier to process.
- Metadata: It includes metadata that can be useful for various NLP tasks.
- Pipeline Integration: It allows different components in the NLP pipeline to work together seamlessly.
Summary
1. Training the NER Model
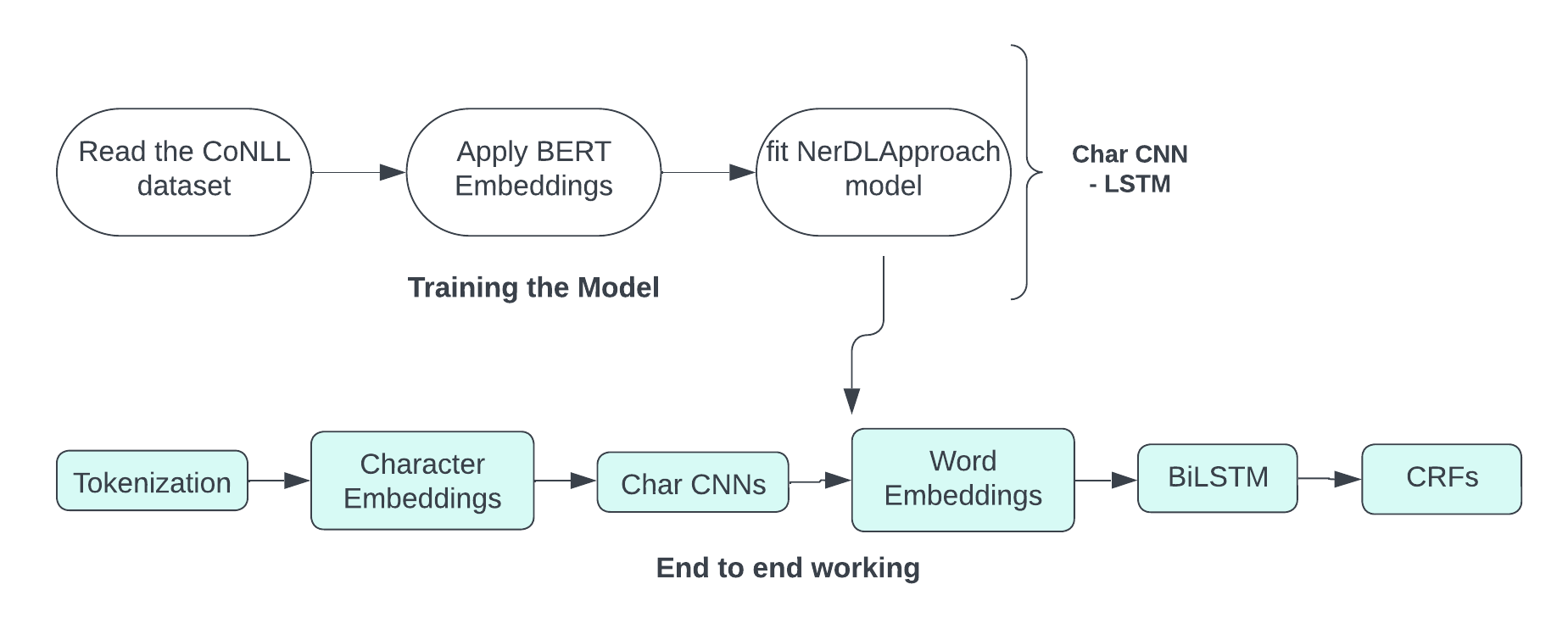
2. OCR and NER Pipeline for Prescription Processing
-
OCR (Optical Character Recognition)
- Converts handwritten doctor prescriptions into machine-readable text using OCR techniques.
-
NER (Named Entity Recognition)
- Input: Text from the OCR step containing unstructured data such as medicine names and dosage instructions.
- BERT Embedding: Converts the input text into context-aware embeddings.
- CHAR CNN-BiLSTM:
- Character-level CNN captures morphological features of words.
- BiLSTM captures bidirectional context of the text sequence.
- CRF (Conditional Random Field): Ensures valid label sequences for structured output like medicine names, dosages, and eating schedules.
Output: Structured data with medicine names, dosages, and schedules extracted from the text.
Actual Output
Handwritten Doctor’s Prescription

Output

Conclusion
In this blog, we explored a comprehensive pipeline for extracting information from handwritten medical prescriptions using AWS Textract and Spark NLP. Starting with the OCR capabilities of AWS Textract, we efficiently transformed handwritten text into machine-readable format, setting the stage for the subsequent NER analysis.
As we concluded with predictions, the seamless interaction of components within the prediction pipeline demonstrated the power of Spark NLP for real-time NER tasks. This approach not only streamlines the processing of medical prescriptions but also holds potential for further applications in healthcare, where accurate data extraction is paramount.
By combining cutting-edge technology with thoughtful design, we can significantly enhance the efficiency of healthcare processes, ultimately leading to improved patient outcomes and better management of medical information. As we continue to innovate in this space, the opportunities for developing advanced applications in the realm of healthcare data processing are endless.
References
[1] “Spark NLP 5.5.1 ScalaDoc - com.johnsnowlabs.nlp.annotators.ner.dl.NerDLApproach,” Sparknlp.org, 2024. https://sparknlp.org/api/com/johnsnowlabs/nlp/annotators/ner/dl/NerDLApproach.
[2] J. Barnard, “What are Word Embeddings? | IBM,” www.ibm.com, Jan. 23, 2024. https://www.ibm.com/topics/word-embeddings
[3] Contextualized Embeddings and Bidirectional Attention: Devlin, J., Chang, M.-W., Lee, K., & Toutanova, K. (2019). BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. Proceedings of NAACL-HLT, 4171–4186. https://arxiv.org/abs/1810.04805.
[4] Handling Out-of-Vocabulary Words and Subword Tokenization: Sennrich, R., Haddow, B., & Birch, A. (2016). Neural Machine Translation of Rare Words with Subword Units. Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), 1715–1725. https://arxiv.org/abs/1508.07909.
[5] Fine-Tuning and Performance on NER: Akbik, A., Blythe, D., & Vollgraf, R. (2018). Contextual String Embeddings for Sequence Labeling. Proceedings of the 27th International Conference on Computational Linguistics, 1638–1649. https://www.aclweb.org/anthology/C18-1139/.
[6] BERT’s Performance in Domain-Specific Applications: Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., & Kang, J. (2020). BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics, 36(4), 1234-1240. https://academic.oup.com/bioinformatics/article/36/4/1234/5566506.
[7] J. P. C. Chiu and E. Nichols, “Named Entity Recognition with Bidirectional LSTM-CNNs,” Transactions of the Association for Computational Linguistics, vol. 4, pp. 357–370, Dec. 2016, doi: https://doi.org/10.1162/tacl_a_00104
[8] “Papers with Code - CoNLL-2003 Dataset,” paperswithcode.com. https://paperswithcode.com/dataset/conll-2003
[9] “Papers with Code - 2010 i2b2/VA Dataset,” Paperswithcode.com, 2022. https://paperswithcode.com/dataset/2010-i2b2-va.
[10] “Spark NLP 5.5.1 ScalaDoc - com.johnsnowlabs.nlp.DocumentAssembler,” Sparknlp.org, 2024. https://sparknlp.org/api/com/johnsnowlabs/nlp/DocumentAssembler.
#Spark-NLP #Handwritten-Prescription-Processing #Medical-Text-Extraction #NER #LSTM #Document #Char-CNN #BERT #BERT-Embeddings #CoNLL